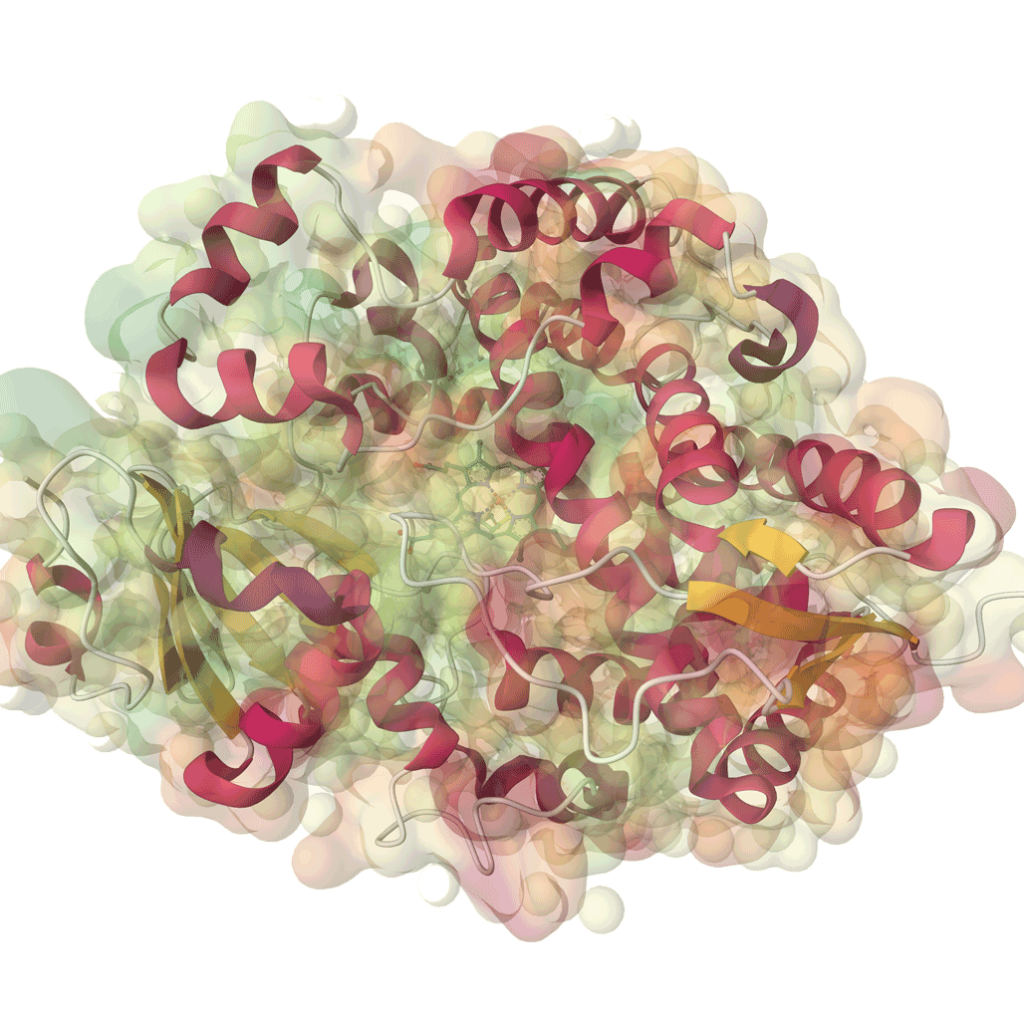
In the ever-evolving landscape of life sciences, understanding “what is molecular dynamics simulations used for” is crucial for advancing research and development. Molecular dynamics simulations (MD) offer profound insights into the behavior of molecules over time, allowing us to study their movements and interactions at an atomic level. This technology plays a pivotal role in various applications, from drug discovery to protein design, and it equips researchers with the tools necessary to predict how biological macromolecules will behave in real-world environments. As we delve into the capabilities of MD simulations, we uncover how they can optimize workflows and enhance our understanding of complex biological systems.
Company Overview
We, at NeoTrident, are committed to revolutionizing the life sciences sector by providing cutting-edge solutions that integrate artificial intelligence with advanced molecular simulation technologies. Our mission is to facilitate groundbreaking discoveries by simplifying the complexities of research workflows. With our latest platform, MaXFlow in Life Science, we bridge the gap between computational chemistry and artificial intelligence, streamlining the process of drug discovery and protein engineering.
Transforming Drug Discovery and Protein Engineering
So, what is molecular dynamics simulations used for in the context of MaXFlow? By integrating Computer-Aided Drug Design (CADD) with AI-Driven Insights (AIDD), MaXFlow allows researchers to conduct advanced Free Energy Perturbation (FEP) simulations. This integration enables us to analyze the thermodynamics of molecular interactions, leading to more informed decisions during the drug development process. The platform provides robust tools for predicting Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET), which are critical factors in assessing the viability of potential drug candidates.
Furthermore, the application of molecular dynamics simulations within MaXFlow is not limited to drug discovery. It also extends to antibody design and enzyme optimization. By employing machine learning algorithms alongside MD simulations, we can achieve rapid and accurate predictions of molecular structures and functionalities. This innovative approach significantly accelerates the protein design process, enabling us to develop new therapeutic antibodies with enhanced specificity and efficacy.
Conclusion
In conclusion, grasping what molecular dynamics simulations are used for is essential for any researcher aiming to stay at the forefront of life sciences innovation. At NeoTrident, we believe that the future of drug discovery and protein engineering lies in our ability to harness these advanced simulations effectively. With MaXFlow in Life Science, we are redefining the standards of bioengineering precision and efficiency. We invite you to explore how our platform can be instrumental in your research efforts, transforming challenges into opportunities for success in the realm of life sciences.
READ ALSO:
https://www.neotridentglobal.com/chemical-compound-drawing-software/
https://www.neotridentglobal.com/molecular-dynamics-simulation-for-all/
https://www.neotridentglobal.com/visual-molecular-dynamics-vmd-software/
https://www.neotridentglobal.com/molecular-dynamics-simulation-protein/
https://www.neotridentglobal.com/best-eln-software/