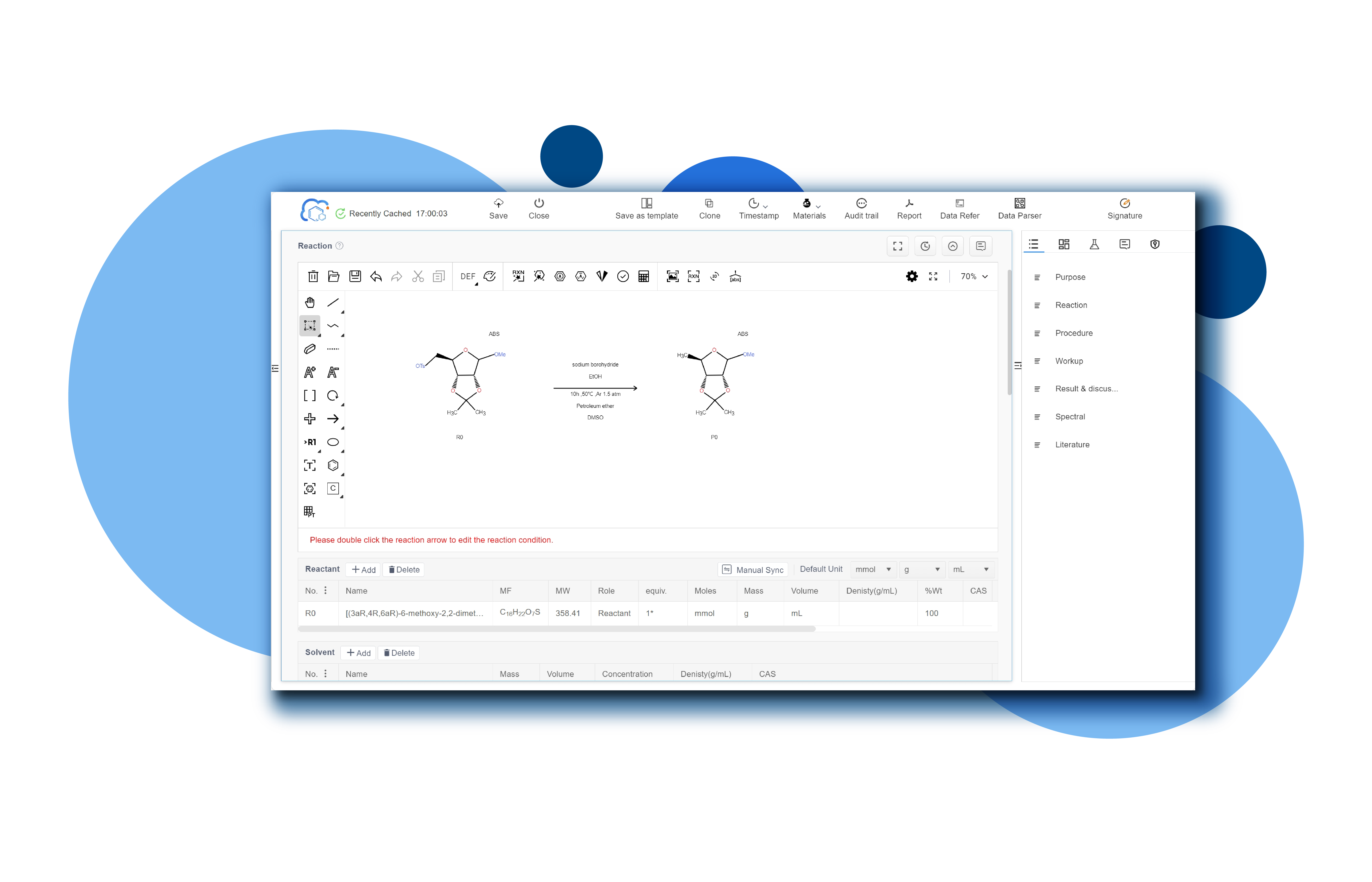
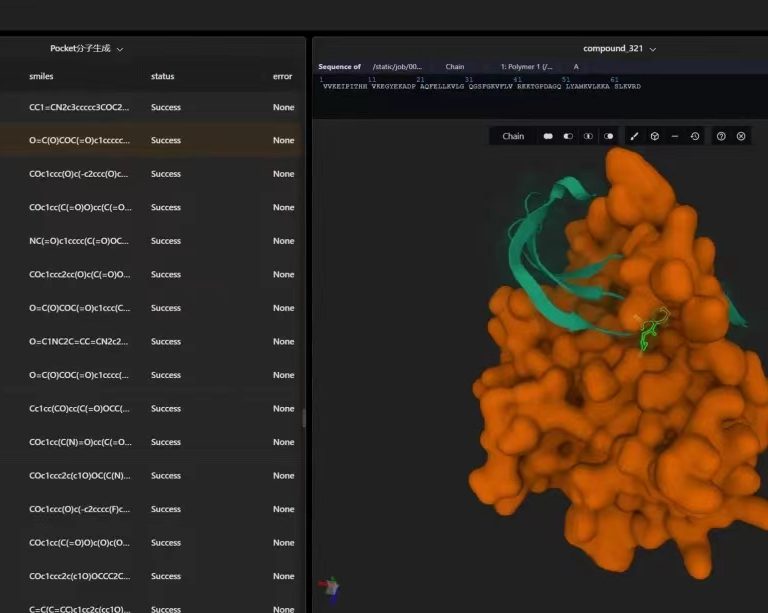
Founded on July 08, 2020, MediLink Therapeutics is an innovative drug development company focusing on antibody-drug conjugates (ADC) and related technologies. MediLink has developed the latest generation of Tumor Microenvironment Activable Linker-payload (TMALIN®) novel ADC platform technology with independent intellectual property rights, which can achieve high DAR value homogeneity with stable coupling. Further improve the therapeutic window of ADC drugs and enhance the therapeutic effect of ADC drugs against tumors, aiming to bring better treatment plans for cancer patients around the world.