With the rise of electronic lab notebooks, molecular biologists and chemists are now able to collaborate more efficiently, streamlining the development of new drugs through standardized workflows. However, as the life sciences industry continues to evolve, there are several challenges that researchers face when working with increasingly complex data and processes in bio-new drug development. One of the main challenges is the need for specialized software solutions that can handle the growing demands of sequence analysis, data management, and experimental reproducibility.
Data Complexity
As drug development processes become more sophisticated, the amount of data generated from biological experiments has increased significantly. Managing and analyzing this data efficiently is crucial for timely and accurate results. Complex workflows, such as gene editing, protein synthesis, and antibody development, require software tools that can handle large datasets, perform detailed analyses, and ensure data traceability across multiple experiments and teams.
R&D Costs and Efficiency
Developing new drugs requires substantial investment in research and development. For laboratories and pharmaceutical companies, optimizing the efficiency of R&D processes is critical to reducing costs and shortening development timelines. Time-consuming tasks, such as manually curating sequences, calculating biochemical properties, and designing primers, can slow down research and lead to errors. Streamlining these processes with integrated tools is essential to improve productivity and ensure high-quality outcomes.
Some researchers have tried using a variety of standalone tools to manage data, but this approach often leads to fragmented workflows, data silos, and inconsistent results, ultimately wasting valuable time and resources.
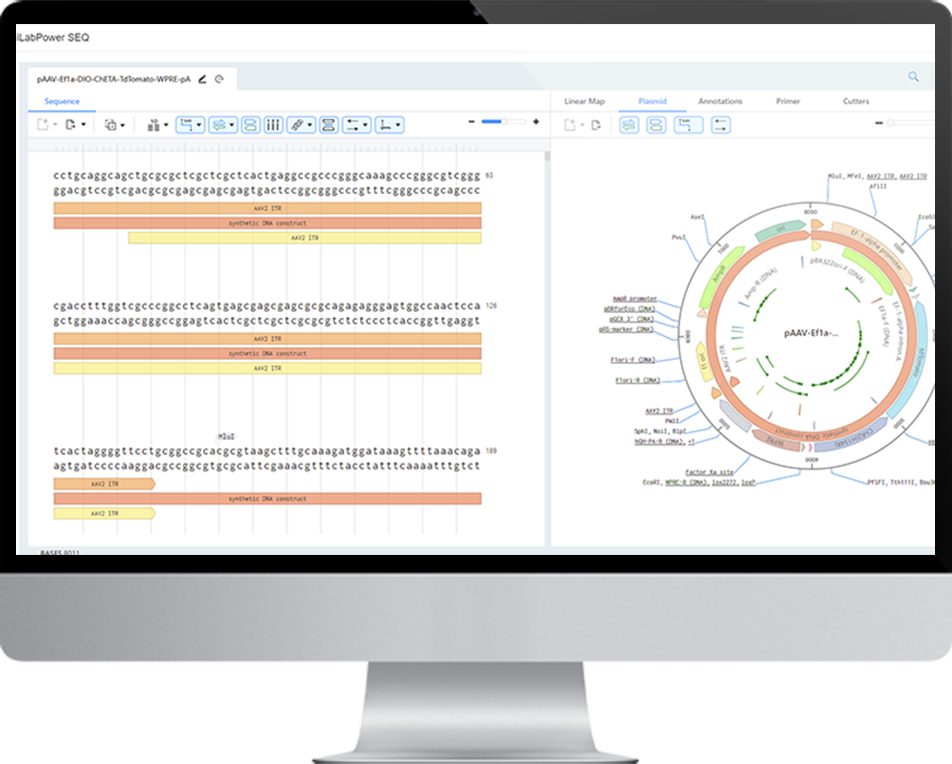
To address these challenges, NeoTrident offers a solution: the iLabPower SEQ Sequence Editor. This tool, part of the iLabPower R&D management platform, provides researchers with an integrated and efficient platform for biological sequence design and analysis. Equipped with visual sequence editing functionality, the tool also allows researchers to document the design process seamlessly in the electronic lab notebook, ensuring full traceability and reproducibility.
iLabPower SEQ Sequence Editor Overview
The iLabPower SEQ Sequence Editor enables users to manage the entire sequence design process—from sequence creation and editing to validation—all within a single platform. For example, in plasmid construction, a common molecular biology experiment, essential steps such as gene and vector selection, PCR primer design, pre-assembly cutting with appropriate endonucleases, and verification via sequencing results can all be conducted within the Sequence Editor. By consolidating these workflows, the platform ensures traceability and reproducibility, which are essential for reliable research. Moreover, applications like CRISPR and circular RNA construction follow similar workflows, making iLabPower SEQ Sequence Editor versatile for a range of bio-design processes.
Key Features of iLabPower SEQ Sequence Editor
- Sequence Creation: The editor supports various ways to create sequences:
- Import FASTA, GenBank, and other annotated sequence files.
- Customize sequence labeling for efficient management and searching.
- Automatically process sequences, including case conversion, coordinate generation, and sequence editing operations, saving time and reducing manual errors.
- Visual Sequence Display: The editor allows users to visualize sequences as linear or circular maps, displaying annotations, primers, and restriction sites in a personalized format. This makes it particularly useful for designing structures such as plasmids, CRISPR, and miRNA.
- Nucleic Acid Sequence Analysis:
- DNA Property Calculation and Primer Design:
Automatically calculates properties like annealing temperature, GC content, and sequence length for primer design. The built-in Primer3 tool generates primer pairs based on user-specified parameters. - Enzyme Site Display:
Identifies restriction enzyme sites using the NEB-Enzyme Finder and displays them for faster sequence editing. - Open Reading Frame (ORF) Recognition:
The editor automatically identifies ORFs, supporting gene expression analysis, which is crucial for drug development.
- DNA Property Calculation and Primer Design:
- Amino Acid Sequence Analysis:
- Protein Biochemical Characterization:
Automatically translates DNA sequences into amino acids, enabling analysis of protein properties, including hydrophobic regions, isoelectric points, and molecular weight. It also facilitates antibody design by identifying CDR (Complementarity-Determining Region) regions critical for antigen recognition.
- Protein Biochemical Characterization:
- Comparative Analysis:
- BLAST:
Provides one-click submission of sequence fragments to NCBI’s BLAST tool for functional prediction. - Dual Sequence Alignment:
Supports alignment of DNA or amino acid sequences, helping users visualize conserved regions and sequence variations.
- BLAST:

iLabPower SEQ Sequence Editor in Bio-New Drug Development
New biopharmaceuticals have shown tremendous promise in treating diseases once deemed incurable, such as cancer and genetic disorders. With its intuitive, efficient, and secure design, iLabPower SEQ Sequence Editor offers a new approach to innovative research in laboratories and pharmaceutical companies. From upstream design to downstream validation, all sequence-related tasks can be conducted on a single platform with robust data security and reproducibility.
Moreover, NeoTrident is committed to continually upgrading the software, improving user interaction, and expanding its bioinformatics capabilities to better serve the bio-new drug R&D landscape. By addressing the unique needs of researchers, NeoTrident aims to support intellectual property protection and accelerate the pace of innovation in biopharmaceuticals.